Chen Lab
Shiming Chen, PhD, Professor, Ophthalmology and Visual Sciences Professor, Developmental Biology

Our Mission
- Decipher the genetic and epigenomic mechanisms regulating photoreceptor gene expression, development and maintenance.
- Understand pathogenesis underlying blinding disease linked to mutations in photoreceptor transcription factors, particularly the Cone-Rod-Homeobox protein CRX.
- Develop mechanism-based therapy strategies for treating CRX-associated photoreceptor disease.
Our Approach
Rod and cone photoreceptors are light-sensing neurons essential for our vision. Their development and maintenance require appropriate expression of a set of specific genes while silencing others. Over-expression and under-expression of certain genes, such as the visual pigment Rhodopsin, can lead to photoreceptor developmental defects or degeneration.
The major goal of our research is to elucidate the molecular mechanisms controlling the expression of photoreceptor genes and how genetic mutations cause gene mis-regulation and defects impacting photoreceptor cell biology and survival. We and others have identified a network of photoreceptor-specific transcription factors (TF) acting along with CRX and chromatin modulators that are essential for regulating target gene expression by binding to specific genomic sites called cis–regulatory elements (CREs), found in gene promoters, enhancers and other sites.
Our current research focuses on 1) deciphering the regulatory grammar of CRX-CRE interactions and the output that determines where, when and how much each photoreceptor gene is expressed; 2) understanding how mutations in either CRX or CREs disrupt the normal regulation and cause photoreceptor diseases. In vivo and ex vivo functional genomics and high-throughput analyses are being used. Examples are listed below:
Omics
- RNAseq transcriptome analyses from bulk cell populations to single cells.
- ChIPseq targetome analyses for TF binding and histone modifications.
- ATACseq for chromatin DNA accessibility.
- 4Cseq interactome combined with DNA & RNA FISH to study enhancer-promoter contacts and genomic organization in the nucleus.
High-throughput
- CREseq or massively-parallel-reporter-assays (MPRAs) for functional CREs in the genome, particularly those bound by CRX.
- CRISPR-Cas9/gRNA mediated genome-wide screens for CREs essential for photoreceptor cell fate.
Disease models
- Generate mouse models for CRX-linked diseases and investigate the pathogenic mechanisms at morphological, electrophysiological and molecular levels.
- Establish human retinal organoid models (miniature retina in a dish) from patient blood cells to investigate the impact of CRX mutations on photoreceptor development and influences of genetic makeup.
- Develop and test new strategies to correct visual defects caused by human CRX mutations.
Research Background
Rods and cones are special neurons carrying out phototransduction that converts the light signal into neuronal signal. Rods are responsible for night vision, while cones are responsible for color vision and visual acuity. Both have a unique structure called Outer Segment (OS) where opsins, the visual pigments, are localized (Fig 1).
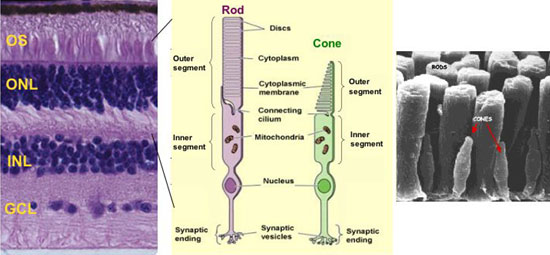
Fig 1: Rods and Cones in mammalian retina. A) A human retinal section showing three neuronal cell layers: outer nuclear layer (ONL) containing the nucleus of rods and cones; inner nuclear layer (INL) containing the nucleus of bipolar, horizontal and amacrine and Muller glial cells; ganglion cell layer (GCL). B) Diagram of rod and cone structure. C) Scanning EM showing the outer segments.
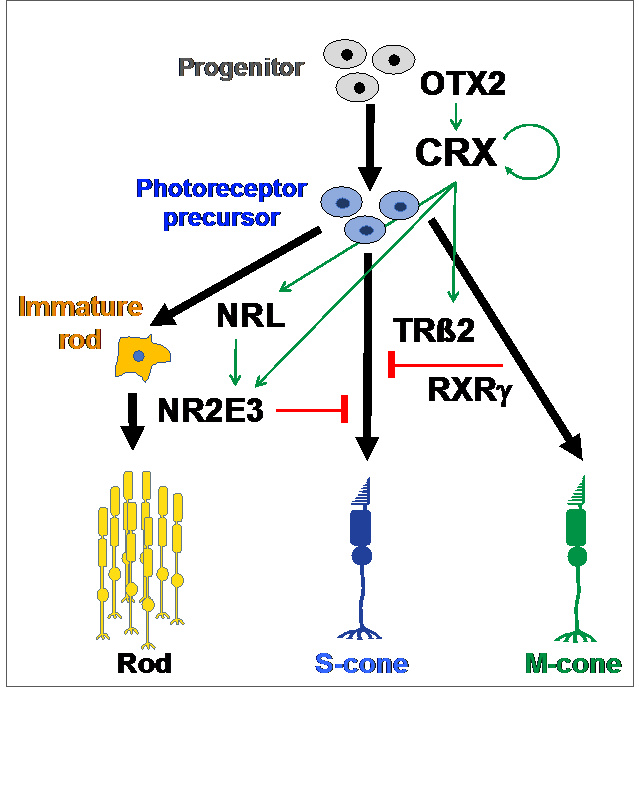
The genetic program controlling rod/cone cell fate is referred as photoreceptor gene-regulatory-network (GRN), built on a set of specific transcription factors (TFs) (Fig 2, left panel) that bind to non-coding DNA regions so called cis-regulatory elements (CREs) (Fig 2, right panel) to determine where, when and how much each gene is expressed.
Key transcription factors (TFs) ➨ Cis-Regulatory Elements (CREs)
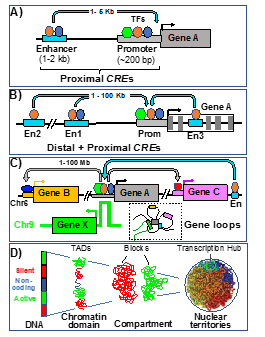
Fig 2: Photoreceptor Gene-Regulatory-Network (GRN). Left: During development, key rod/cone TFs are activated in CRX-expressing cells (photoreceptor precursor) and act with CRX to specify rod versus cone cell fate. Right: Each target gene is regulated by interactions among TF-bound cis-regulatory elements (CREs). The chromatin containing these CREs/genes undergoes multiple levels of genomic organization in the nucleus as modeled in A-D: By forming gene loops, the promoter of a gene interacts with its proximal enhancer (A), distal enhancers (B), and additional long-range enhancers (C). The whole genome in the nucleus is further organized into distinct domains and active/silent compartments and territories (D).
CRX is an otd/OTX-like homeodomain TF predominantly expressed in rods, cones (Fig 3A) and their precursors. CRX is essential for photoreceptor gene expression, development and maintenance. In Crx knockout mice (Crx-/-), rods/cones fail to generate proper structure and function due to gene mis-regulation, leading to “blindness” and retinal degeneration. CRX has a N-terminal DNA-binding homeodomain and C-terminal trans-activation domains (Fig 3B). CRX acts by 1) binding to approximately six thousand CREs in the genome, 2) recruiting co-activators (epigenetic regulators) to a subset of CREs for local chromatin remodeling, and 3) activating transcription of target genes along with other TFs (Fig 3C).
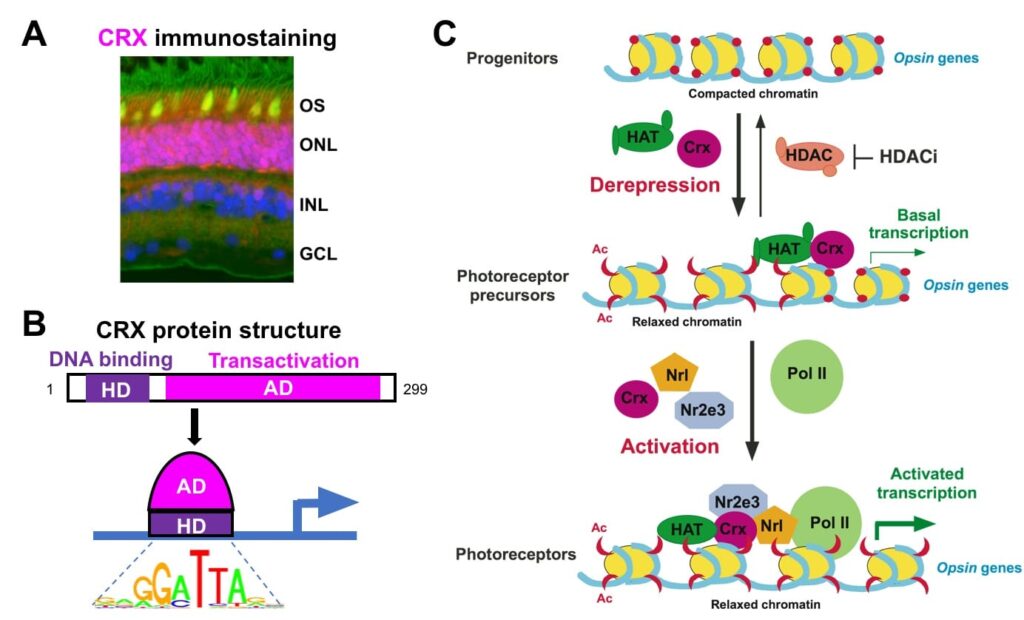
Fig 3: CRX acts in rods/cones to regulate gene expression. (A) Immunostaining of a monkey retinal section, showing CRX (purple) is localized to the rod/cone nucleus (Blue-nucleus, Green-rod OS, Yellow-cone OS). (B) CRX contains two major functional domains and binds to a consensus DNA motif. (C) Model for CRX mechanism of action: CRX recruits epigenetic regulators, such as histone acetyltransferase (HAT), to configure an “open” chromatin at target genes, and activates transcription by interacting with other TFs and the RNA Pol II transcription machinery (adapted from Peng & Chen, 2007). CRX-mediated chromatin remodeling occurs at the level of DNA accessibility, histone modifications and long-range genomic interactions (Peng & Chen 2011; Ruzycki et al, 2018).
Mutations in the human CRX gene are associated with autosomal dominant and de novo photoreceptor diseases varying in age onset and phenotype severity, including Leber congenital amaurosis (LCA), cone-rod dystrophy (CRD) and retinitis pigmentosa (RP). Affected cones and rods show developmental abnormalities and degeneration. No treatment is currently available. Disease-causing mutations distribute along CRX protein and can be divided into four classes based on biochemical characteristics and pathogenic mechanisms of mutant proteins (Fig 4).
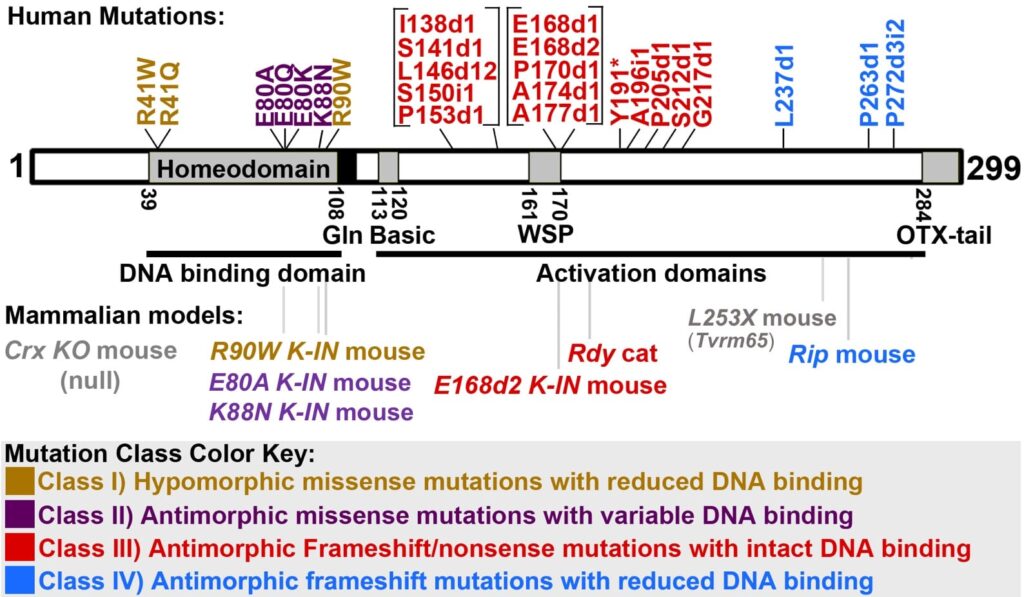
Fig 4: Four classes of disease-causing human CRX mutations and available animal models (adapted from Tran & Chen).
Meet The Team

Shiming Chen, PhD
Dr. Bernard and Janet R. Becker Distinguished Professor in Ophthalmology
- Email: chenshiming@nospam.wustl.edu
- Chen Lab page
- Research Profiles
- Director of the Molecular Genetics Core

Madyson Brown
Postbaccalaureate researcher DCBRM
- Email: bmadyson@wustl.edu
Education:
- B.A. in Psychology and Neuroscience, Penn State University.
Project: Impact of mutant forms of CRX on photoreceptor morphology and function.
Fun Fact: I like to spend time with family and friends, outside, and watch movies.

Sun Chi, Ph.D.
Postdoctoral Research Associate, Instructor
- Phone: 314-747-4351
- Email: sunchi@wustl.edu
Education:
- B. S. in Chemical & Biomolecular Engineering, Nanyang Technological University (NTU), Singapore.
- Ph. D. in Neuroscience, University of Idaho, Idaho.
- Postdoctoral fellow in Chen Lab, WashU
Project: CRX and its cis-regulatory elements in photoreceptor development and maintenance.
Fun fact: I like to play video games and watch football (soccer)!

Isabella Gomes
Research Fellow
- Email: i.gomes@wustl.edu
Education:
-
B.A. in Ecology & Evolutionary Biology (Infectious Diseases), Princeton University
- M.P.H in Epidemiology & Biostatistics, Johns Hopkins Bloomberg School of Public Health
- M.A., Columbia Journalism School
-
MD Candidate, Class of 2026, WashU
Project: Clinical presentations of CRX Retinopathies
Fun Fact: I’ve been an ordained minister for 8 years. I have not married anyone yet.

Maya Holman
Research Tech II
- Email: mayah@wustl.edu
Education:
- B.S. DePaul University, where she studied integrative neuroscience, biology, and entrepreneurship.
Project: Rescuing CRX function via AAV-mediated gene therapy.
Fun Fact: I love creating art and selling my work at an independent bookstore in Chicago.

Max Liu
WashU sophomore, Biology major
- Email: liu.max@wustl.edu
Project: Immunohistochemistry of mouse retinal sections.
Fun fact: I like to cook and write my own recipes.

Mingyan (June) Yang, B.S.
Research Technician II
- Phone: 314-747-4351
- Email: yangm@wustl.edu
Education:
- B.S. Shandong Sport College, Jinan, P. R. China.
Project: Mouse models of retinal degeneration
Fun fact: I have a hobby of grooming my Yorkshire Terrier. Although many people say I’m not good at it, I firmly believe that my dog looks beautiful no matter how I groom him!

Maggie Yu
WashU junior, Major in Biology and Neuroscience
- Email: maggie.m.yu@wustl.edu
Project: Immunohistochemistry of mouse retinal sections.
Fun Fact: My favorite animal is the red panda!

Xiaodong Zhang, Ph.D.
Staff Scientist
- Phone: 314-747-4351
- Email: xzhang26@wustl.edu
Education:
- B.S. Nanjing University, China.
- M.S. University of Louisville, Kentucky.
- Ph.D. University of Louisville, Kentucky.
Project: General transcription factor complexes in photoreceptor gene expression, development and maintenance.
Fun fact: I enjoy gardening in my free time.
Previous Lab Members
Member Name: Yiqiao Zheng, Ph.D. 2018-2024 – Ph.D. student in Molecular Genetics & Genomics
Project: Pathogenic mechanisms of CRX homeodomain mutations
Current Position: Postdoctoral fellow with Dr. Daniel Lew, Biology, MIT
Member Name: Inez Oh, Ph.D. 2016-2019 – Postdoctoral Research Associate
Project: RNA metabolism in normal and diseased retinas
Current Position: Staff Scientist, Institute for Informatics, Washington University
Member Name: Philip A. Ruzycki Ph.D. 2012-2017 – Ph.D. student in Molecular Genetics and Genomics
Project: Epigenetic regulation of photoreceptor gene expression
Current position: Assistant Professor, Ophthalmology & Vision Sciences, Washington University
Member Name: Diana S. Brightman, Ph.D. 2010-2016 – Ph.D. student in Molecular Cell Biology
Project: Histone methyltransferases and demethylases in retinal development.
Current position: Genetic Counselor, Cincinnati Children’s Hospital
Member Name: Nicholas M.A. Tran, Ph.D. 2008-2014 -Ph.D. student in Molecular Genetics & Genomics
Project: The mechanisms by which human CRX mutations cause dominant retinopathies
Current position: Assistant Professor, Molecular and Human Genetics, Baylor College of Medicine
Member Name: Guang-Hua Peng, M.D., Ph.D. 2002-2011 – Senior Staff Scientist
Current position: Professor, Hernan University School of Med, China
Member Name: Xin Fang, B.S. 2017-2018 – Research Technician II
Project: The histone methyltransferases MLL1 and MLL2 in retinal development and maintenance
Current position: Dental student, University of the Pacific Dugoni School of Dentistry
Member Name: Courtney D. Linne, B.A. 2016-2017 – Research Technician II and Undergraduate Research Assistant.
Project: Animal models of CRX-associated blinding diseases.
Current position: M.D./Ph.D. student, University of Cincinnati College of Medicine
Member Name: Hui Wang, M.D. 2006-2013 – Research Technician II
Project: Mouse models of blinding diseases
Member Name: Rachel L. Grant, B.A. 2013-2015 -Undergraduate Research Assistant
Project: The histone methyltransferase MLL1 in retinal development and maintenance
Current position: Ph.D. student in Developmental Biology, Stanford University
Member Name: Ray Suzuki, B.A. 2011-2013 – Undergraduate Research Assistant
Project: The histone methyltransferase MLL1 in retinal development and maintenance
Current position: Technical Consultant, Phenomenex Company
Chen Lab Photo Album
Publications
View Shiming Chen’s NCBI publications on PubMed»
- Aberrant homeodomain-DNA cooperative dimerization underlies distinct developmental defects in two dominant CRX retinopathy models. Zheng Y, Stormo GD, Chen S bioRxiv. 2024 Mar 14;. doi: 10.1101/2024.03.12.584677. PubMed PMID: 38559186; PubMed Central PMCID: PMC10979960.
- Missense mutations in CRX homeodomain cause dominant retinopathies through two distinct mechanisms. Zheng Y, Sun C, Zhang X, Ruzycki PA, Chen S. Elife. 2023 Nov 14;12. doi: 10.7554/eLife.87147. PubMed PMID: 37963072; PubMed Central PMCID: PMC10645426.
- Rho enhancers play unexpectedly minor roles in Rhodopsin transcription and rod cell integrity. Sun C, Ruzycki PA, Chen S. Sci Rep. 2023 Aug 9;13(1):12899. doi: 10.1038/s41598-023-39979-6. PubMed PMID: 37558693; PubMed Central PMCID: PMC10412641.
- Essential Functions of MLL1 and MLL2 in Retinal Development and Cone Cell Maintenance. Sun C, Zhang X, Ruzycki PA, Chen S. Front Cell Dev Biol. 2022;10:829536. doi: 10.3389/fcell.2022.829536. eCollection 2022. PubMed PMID: 35223853; PubMed Central PMCID: PMC8864151.
- MLL1 is essential for retinal neurogenesis and horizontal inner neuron integrity. Brightman DS, Grant RL, Ruzycki PA, Suzuki R, Hennig AK, Chen S. Scientific reports. 2018; 8(1):11902.
- CRX directs photoreceptor differentiation by accelerating chromatin remodeling at specific target sites. Ruzycki PA, Zhang X, Chen S. Epigenetics & chromatin. 2018; 11(1):42.
- Crx-L253X Mutation Produces Dominant Photoreceptor Defects in TVRM65 Mice. Ruzycki PA, Linne CD, Hennig AK, Chen S. Investigative ophthalmology & visual science. 2017; 58(11):4644
- CrxRdy Cat: A Large Animal Model for CRX-Associated Leber Congenital Amaurosis.Occelli LM, Tran NM, Narfström K, Chen S, Petersen-Jones SM. Investigative Ophthalmology & Visual Science. 2016; 57(8):3780-92.
- Nrl-Cre transgenic mouse mediates loxP recombination in developing rod photoreceptors. Brightman DS, Razafsky D, Potter C, Hodzic D, Chen S. Genesis 2016; 54(3):129-35.
- Graded gene expression changes determine phenotype severity in mouse models of CRX-associated retinopathies. Ruzycki PA, Tran NM, Kefalov VJ, Kolesnikov AV, Chen S. Genome Biology. 2015; 16:171.
- The transcription factor GTF2IRD1 regulates the topology and function of photoreceptors by modulating photoreceptor gene expression across the retina. Masuda T, Zhang X, Berlinicke C, Wan J, Yerrabelli A, Conner EA, Kjellstrom S, Bush R, Thorgeirsson SS, Swaroop A, Chen S, Zack DJ. Journal of Neuroscience 2014; 34(46):15356-68.
- Mechanisms of blindness: animal models provide insight into distinct CRX-associated retinopathies. Tran NM, Chen S. Developmental Dynamics. 2014; 243(10):1153-66.
- Mechanistically distinct mouse models for CRX-associated retinopathy. Tran NM, Zhang A, Zhang X, Huecker JB, Hennig AK, Chen S. PLoS Genetics. 2014; 10(2):e1004111.
- Transcription coactivators p300 and CBP are necessary for photoreceptor-specific chromatin organization and gene expression. Hennig AK, Peng GH, Chen S. PLoS One. 2013; 8(7):e69721.
- Active opsin loci adopt intrachromosomal loops that depend on the photoreceptor transcription factor network. Peng GH, Chen S. Proceedings of the National Academy of Sciences of the United States of America. 2011; 108(43):17821-6.
- Pias3-dependent SUMOylation controls mammalian cone photoreceptor differentiation. Onishi A, Peng GH, Chen S, Blackshaw S. Nature Neuroscience. 2010; 13(9):1059-65