This Research Insight covers a recent publication from the Morgan Lab. Here, we highlight the stand-out performance of a modified deep learning-based tool to assess the quality of electron micrographs in real-time.
In their recent paper published in Microscopy and Microanalysis, scientists in the lab of Joshua Morgan, PhD, assistant professor of ophthalmology at WashU Medicine, demonstrate the remarkable performance of a deep learning-based tool to streamline the evaluation of electron micrographs taken of the mouse brain. The high reliability of the Quality Evaluation Network (QEN) demonstrated here supports its adoption by the Morgan Lab to fully automate and improve the quality of electron micrographs acquired for serial section scanning electron microscopy (ssSEM) studies.
Bottlenecks in ssSEM imaging
In ssSEM studies, biological samples are cut into ultrathin sections (10-90 nm thick) and imaged by scanning electron microscopy. The resulting images generated across the depth of the tissue are then stacked together to generate high-resolution, three-dimensional reconstructions of the sample, in which the cellular architecture of the sample is visible and can be annotated. While this technique yields highly informative datasets that can be used to answer a host of biological questions, the generation of these datasets is time-consuming and laborious, as it requires reliable image acquisition and alignment across potentially thousands of sections, depending on sample volume. Tools to automate image acquisition and assess the quality of individual images without manual intervention are therefore key to maximizing the usable output of ssSEM image acquisition pipelines.
Advancing quality assessment beyond current deep learning algorithms

Here, Mahsa Bank-Tavakoli, PhD, a postdoctoral fellow in the Morgan Lab, led an effort to compare the accuracy and reliability of three convolutional neural networks to automatically evaluate the quality of electron micrographs. Bank-Tavakoli used 2000 human-assessed electron micrographs taken of the mouse brain to train and an additional 500 images to test a modified ResNet-50 convolutional neural network – the QEN – to show that it can rapidly and reliably predict image quality. Bank-Tavakoli and Morgan benchmarked their modified QEN against other established neural networks – the DCNN-IQA-14 and MIQA – which are used to quality check ultrasound and other biomedical images, respectively.
Optimizing efficiency for real-time image assessment
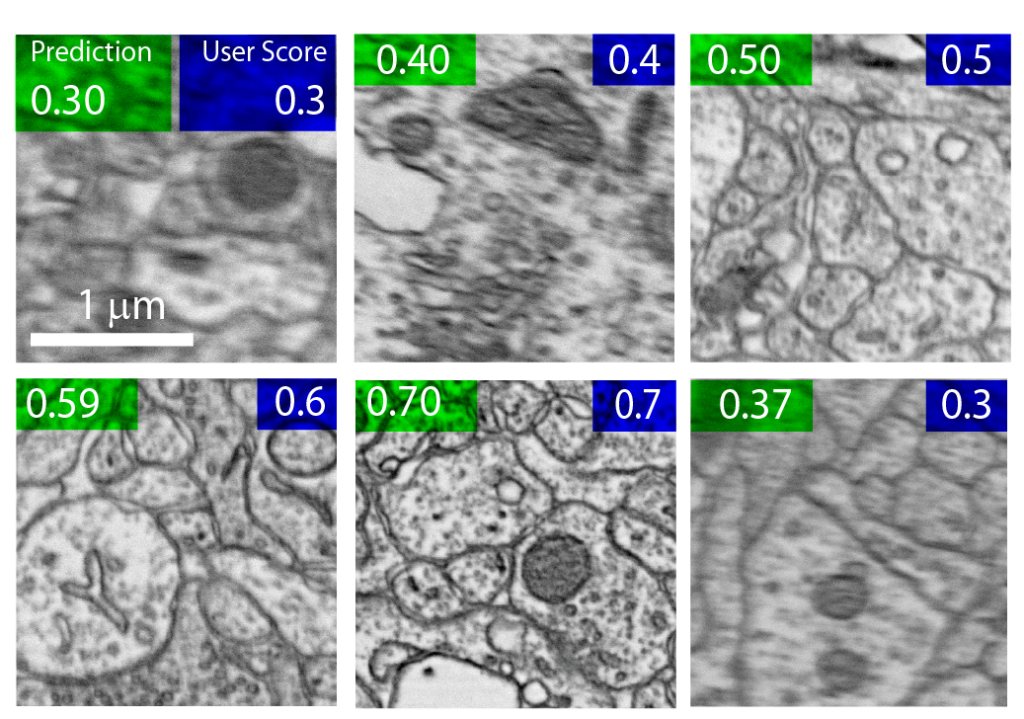
The modified QEN made few errors when predicting the human-assigned quality scores for each image. However, assessing the quality of high-resolution images (20480 x 20480 pixels) in parallel with image acquisition was time-consuming and posed a significant bottleneck to image acquisition speed.
Bank-Tavakoli and Morgan therefore tested the ability of the QEN to use small image patches (512 x 512 pixels) to recapitulate the human-assigned quality scores of their source images. They found that as few as 3 random patches from a single image was sufficient to assess the quality of the high-resolution image rapidly and with high accuracy.
A fully automated image acquisition pipeline
The results of this comparative study supported the Morgan Lab’s adoption of a fully automated image acquisition pipeline for ssSEM studies. The lab now uses the modified QEN to assess the quality of individual electron micrographs during image acquisition to automatically determine which sample images must be reacquired. By assessing the quality of small image patches – rather than full, high-resolution images – the QEN can accurately benchmark image quality in seconds.
The Morgan Lab has published the code for running and training their modified QEN, which may be suitable for other applications that require the analysis of monochrome two-dimensional images of cell membranes and organelles. The code can be downloaded from GitHub.